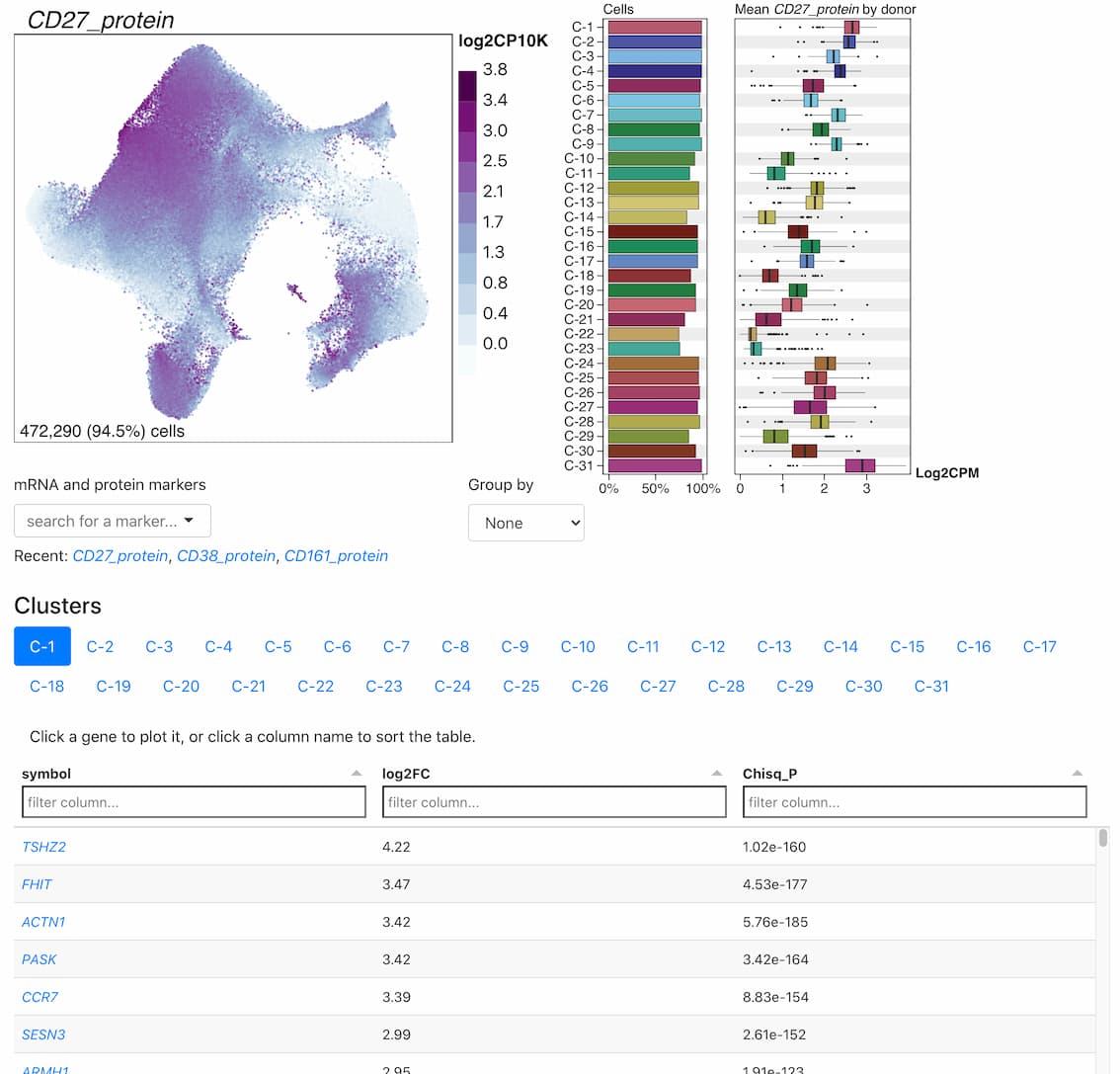
Cell Clusters
Metadata variables, protein expression, and gene expression in two-dimensional embeddings.
Launch Viewer →Multimodally profiling memory T cells from a tuberculosis cohort identifies cell state associations with demographics, environment and disease
Nature Immunology (2021) · doi:10.1038/s41590-021-00933-1
Abstract
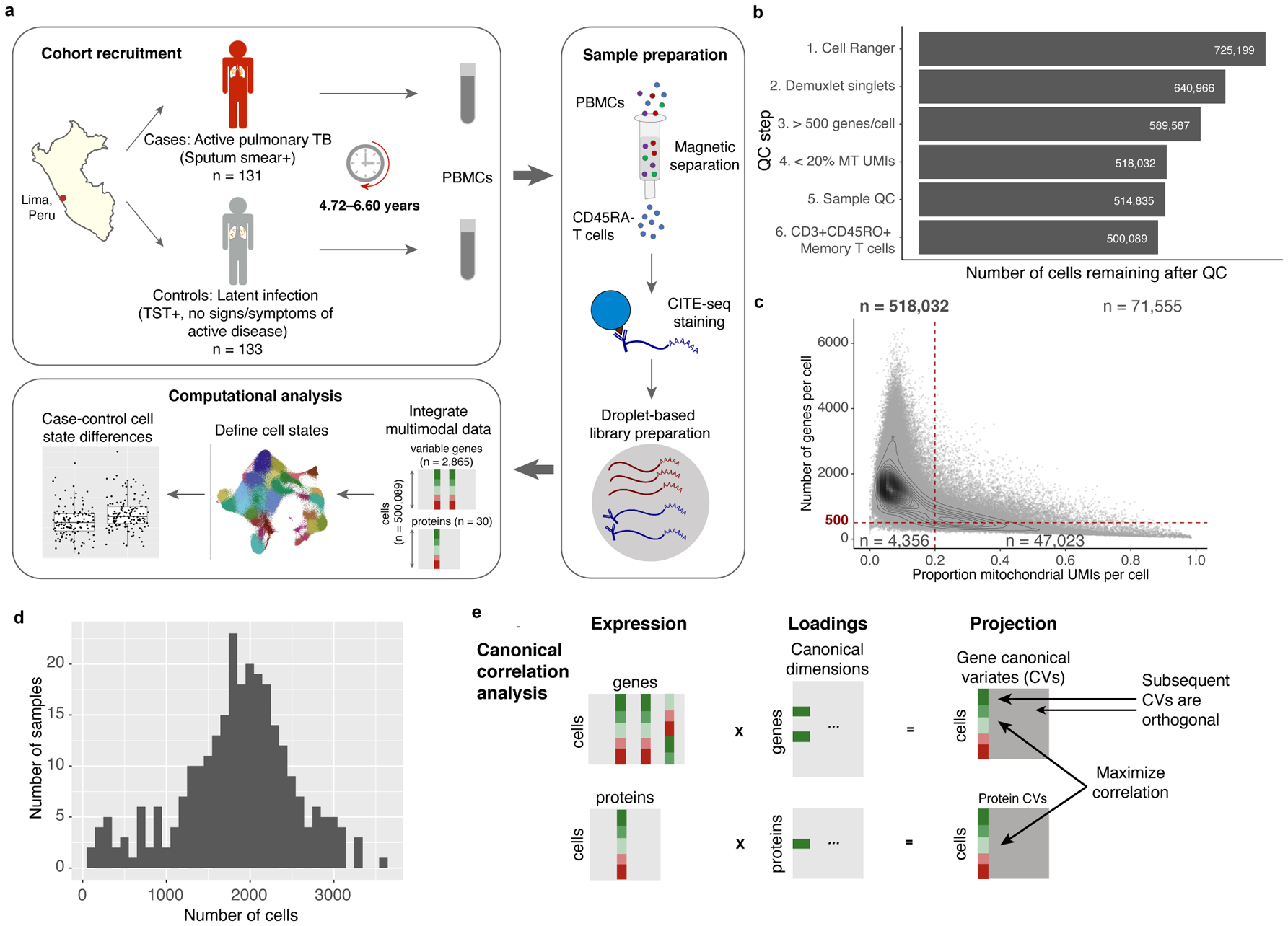
Multimodal T cell profiling can enable more precise characterization of elusive cell states underlying disease. Here, we integrated single-cell RNA and surface protein data from 500,089 memory T cells to define 31 cell states from 259 individuals in a Peruvian tuberculosis (TB) progression cohort. At immune steady state >4 years after infection and disease resolution, we found that, after accounting for significant effects of age, sex, season and genetic ancestry on T cell composition, a polyfunctional type 17 helper T (TH17) cell-like effector state was reduced in abundance and function in individuals who previously progressed from Mycobacterium tuberculosis (M.tb) infection to active TB disease. These cells are capable of responding to M.tb peptides. Deconvoluting this state-uniquely identifiable with multimodal analysis-from public data demonstrated that its depletion may precede and persist beyond active disease. Our study demonstrates the power of integrative multimodal single-cell profiling to define cell states relevant to disease and other traits.
Resources
Download Data
NCBI GEO: GSE158769
Source Code
Analysis: immunogenomics/TB_Tcell_CITEseq
Contact
Questions or comments? Get in touch.