Explore gene expression along the T cell innateness gradient
Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions
Maria Gutierrez-Arcelus, Nikola Teslovich, Alex R. Mola, Rafael B. Polidoro, Aparna Nathan, Hyun Kim, Susan Hannes, Kamil Slowikowski, Gerald F. M. Watts, Ilya Korsunsky, Michael B. Brenner, Soumya Raychaudhuri, Patrick J. Brennan.
Nature Communications 2019. doi: 10.1038/s41467-019-08604-4
Abstract
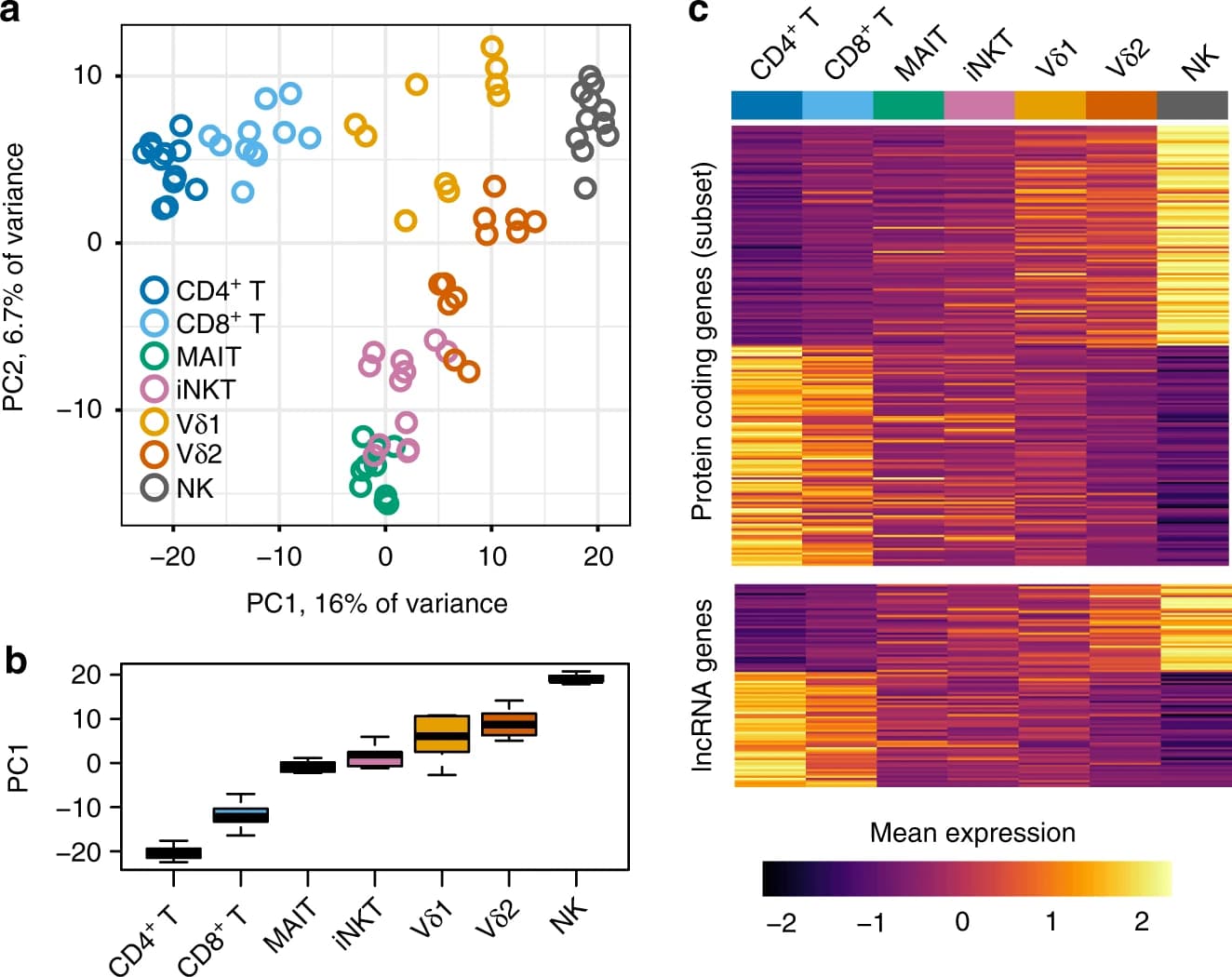
How innate T cells (ITC), including invariant natural killer T (iNKT) cells, mucosal-associated invariant T (MAIT) cells, and γδ T cells, maintain a poised effector state has been unclear. Here we address this question using low-input and single-cell RNA-seq of human lymphocyte populations. Unbiased transcriptomic analyses uncover a continuous ‘innateness gradient’, with adaptive T cells at one end, followed by MAIT, iNKT, γδ T and natural killer cells at the other end. Single-cell RNA-seq reveals four broad states of innateness, and heterogeneity within canonical innate and adaptive populations. Transcriptional and functional data show that innateness is characterized by pre-formed mRNA encoding effector functions, but impaired proliferation marked by decreased baseline expression of ribosomal genes. Together, our data shed new light on the poised state of ITC, in which innateness is defined by a transcriptionally-orchestrated trade-off between rapid cell growth and rapid effector function.
View the data
On this website, we provide an interactive data browser to view association statistics for each gene with the innateness gradient. We also show a summary of the expression data, and correlated genes to the selected gene.

Contact us
Please contact Dr. Maria Gutierrez-Arcelus with any questions, requests, or comments.
Get the data
The data presented here comes from the laboratories of:
Download the data from NCBI GEO:
Get the code
Get the code for the analysis:
Get the code for this website:

