Looking for the AMP RA Phase I data (SDY998)? You're in the right place.
ImmPort is the official repository for this dataset, but downloading files requires a few setup steps. This guide walks you through the entire process—from creating an account to loading data in R.
Step 1: Find the study on ImmPort
Start by visiting the ImmPort study page for SDY998:
You can also find this link on the AMP RA Phase I page or the GitHub repository.
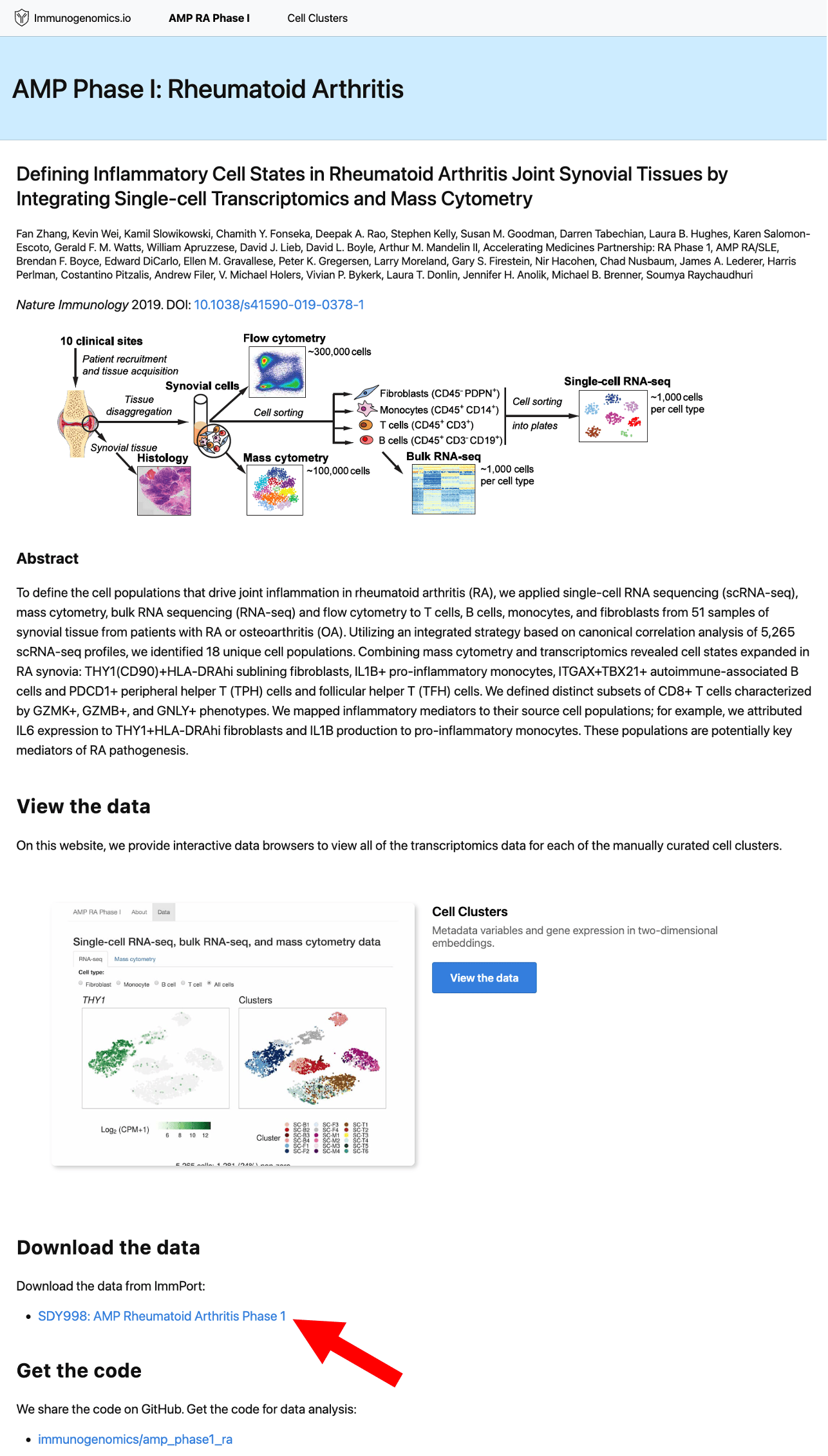
Step 2: Create an ImmPort account
To download files, you’ll need a free ImmPort account. Click Study Package (Web) in the top navigation under “Download Options”.
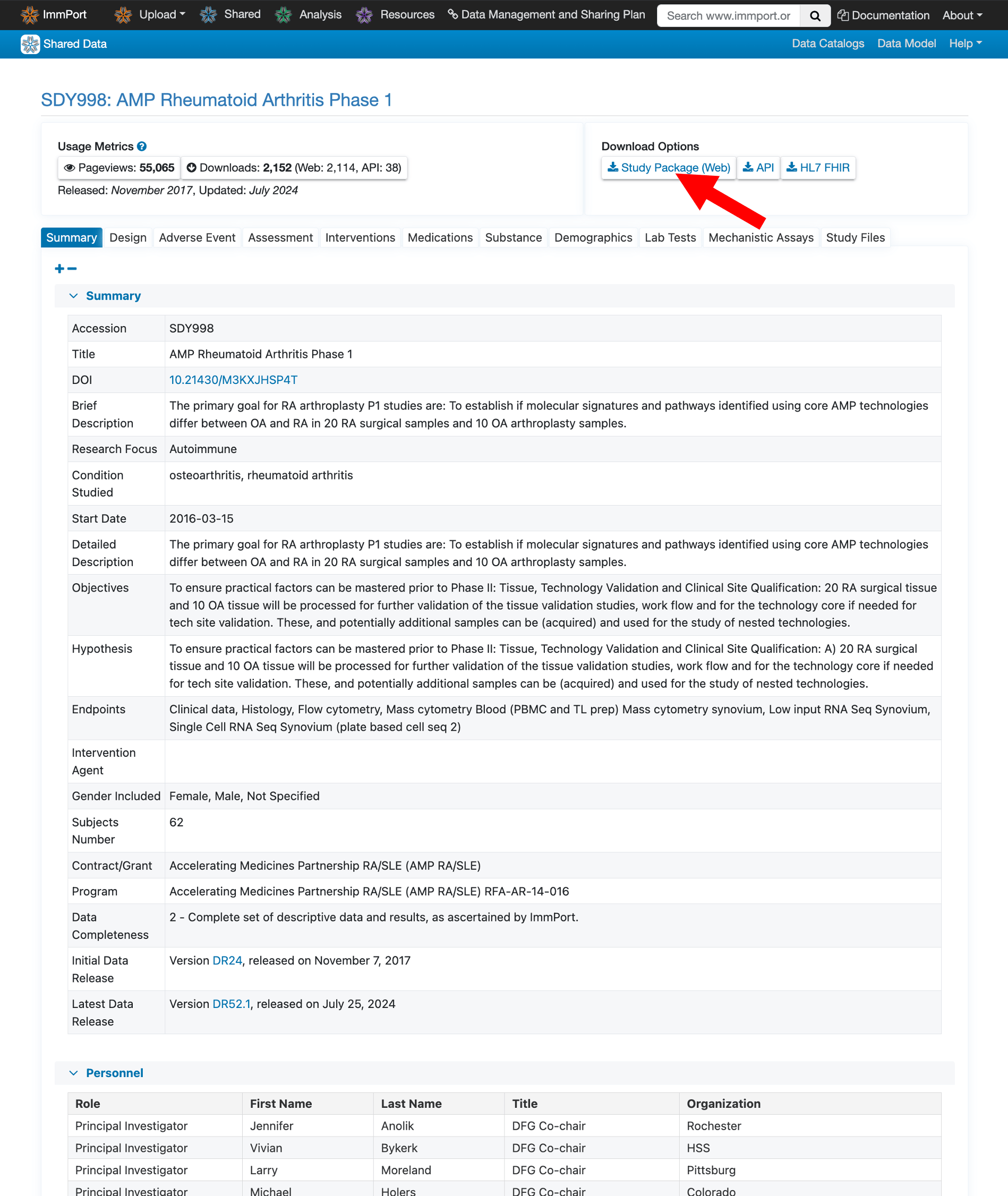
Since downloads require registration, you will be redirected to a login and registration page. Now click Register and fill out the registration form with your details.

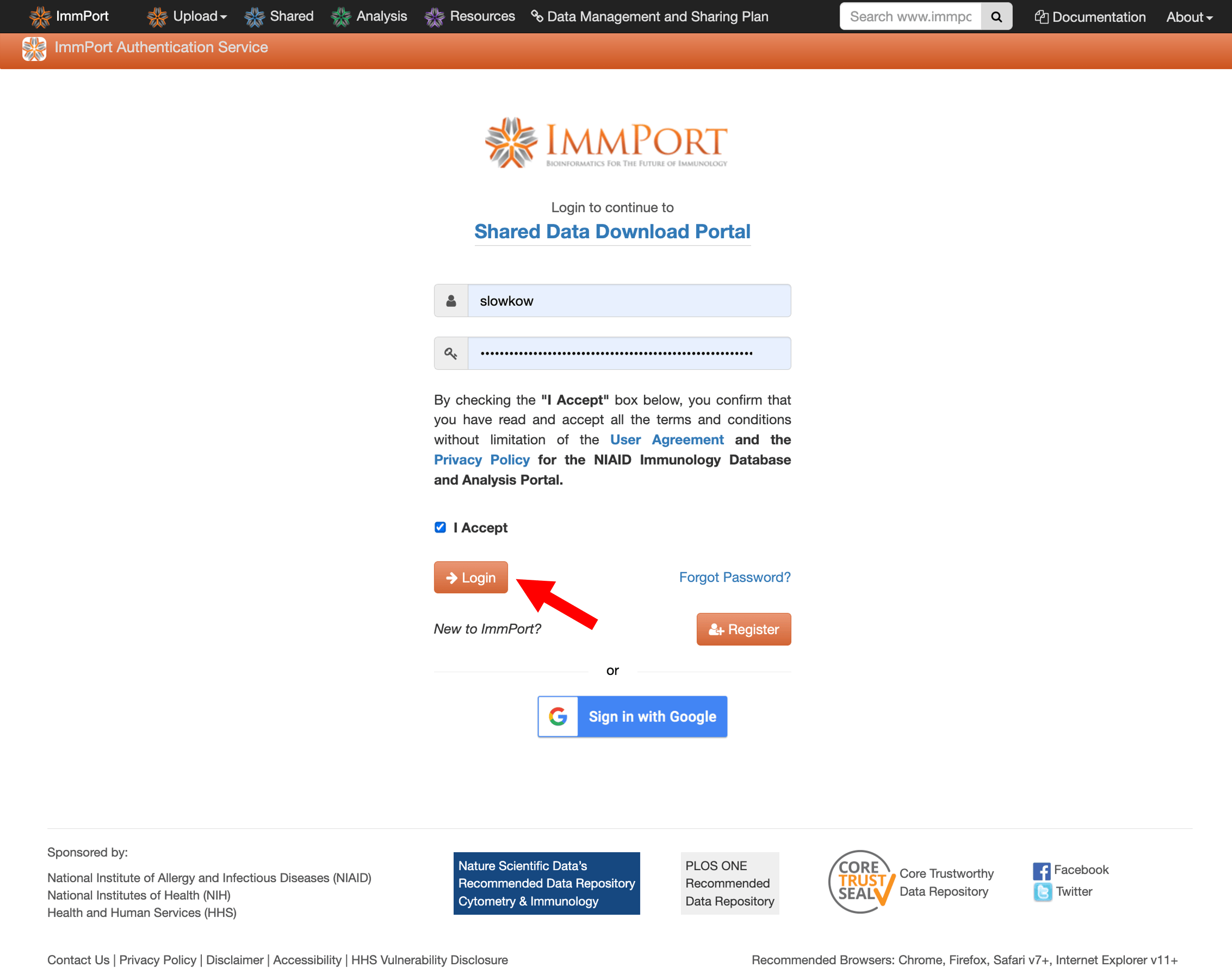
Step 3: Log in to ImmPort
Once your account is confirmed, log in using your credentials.
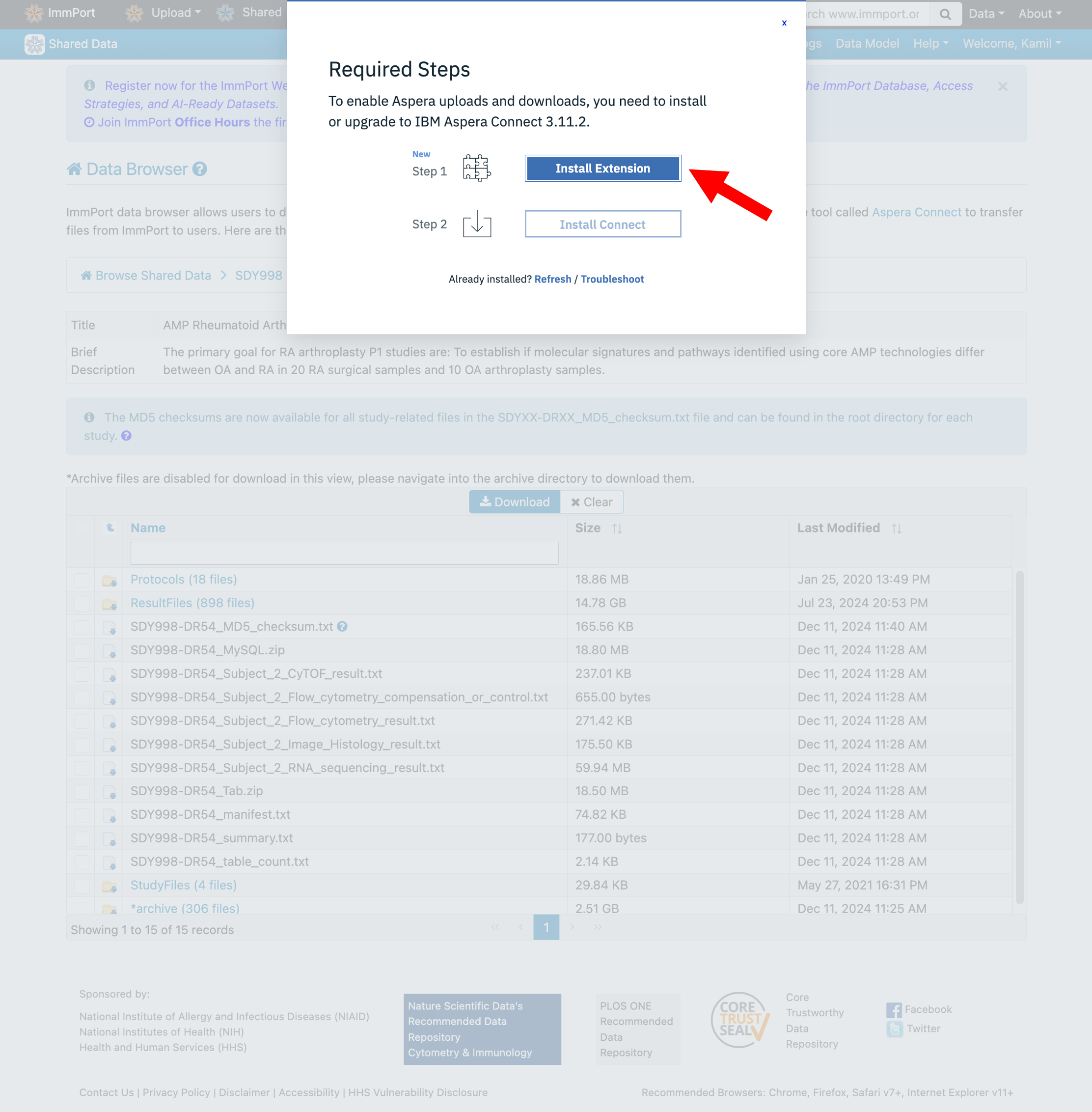
Step 4: Install Aspera Connect
ImmPort uses IBM Aspera Connect for fast, reliable file transfers. You’ll need to install both the browser extension and desktop application.
Step 5: Download and read the README
Before downloading data files, take a moment to understand what’s available. Navigate to:
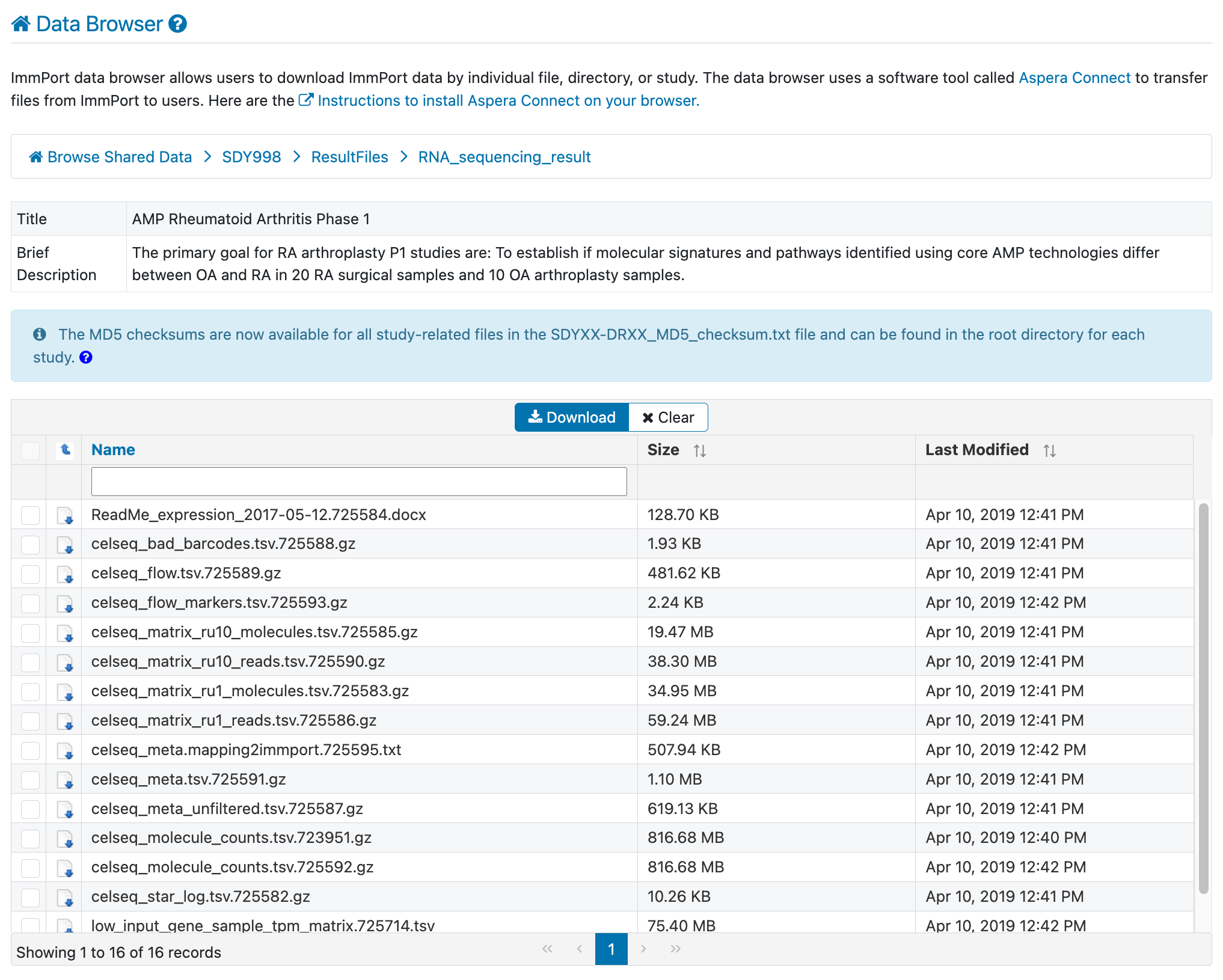
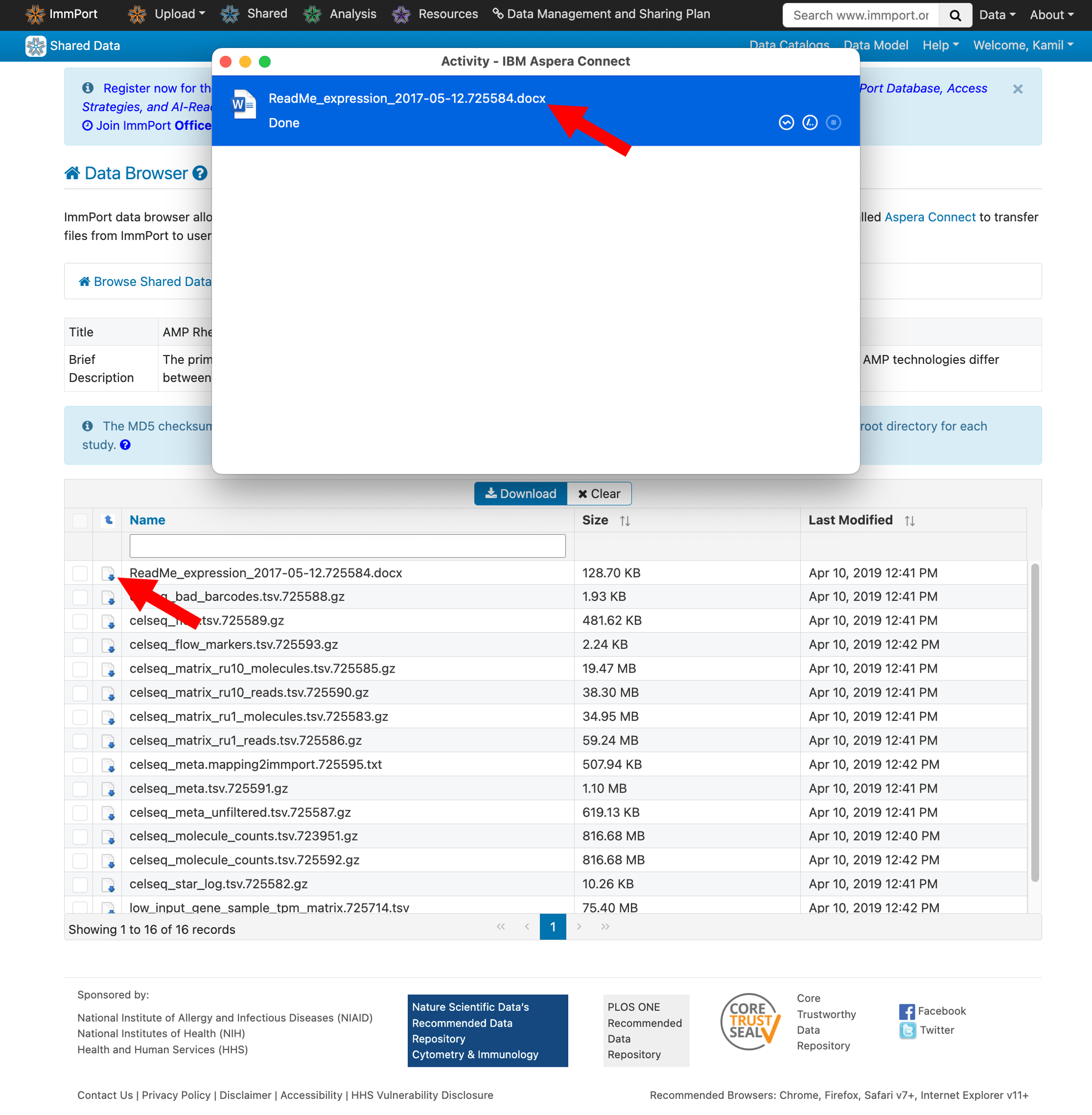
Download the README file first—it describes each file and helps you identify exactly what you need.
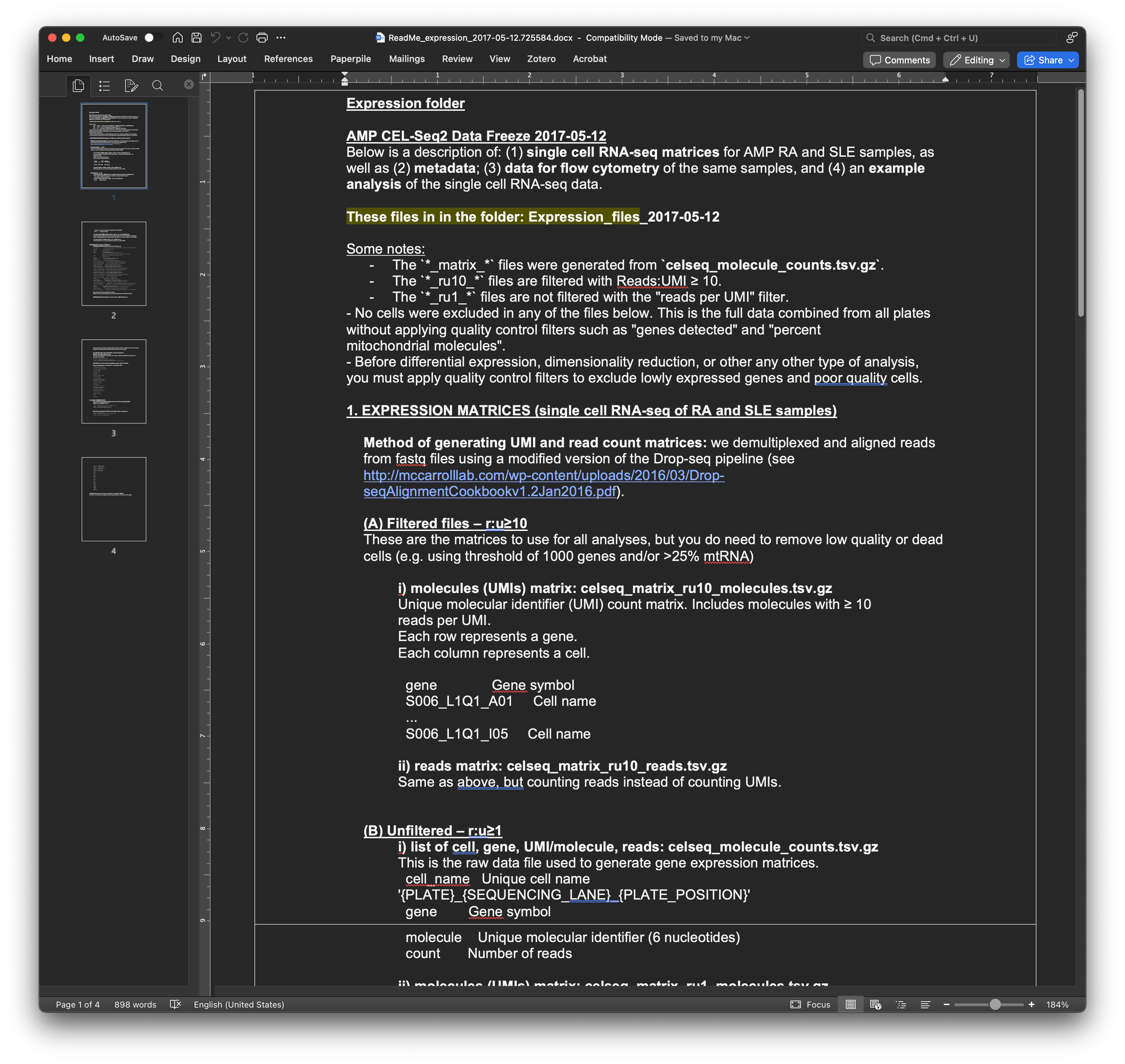
Step 6: Download the data files
Select the files you need for your analysis and click to download. Aspera Connect will handle the transfer.
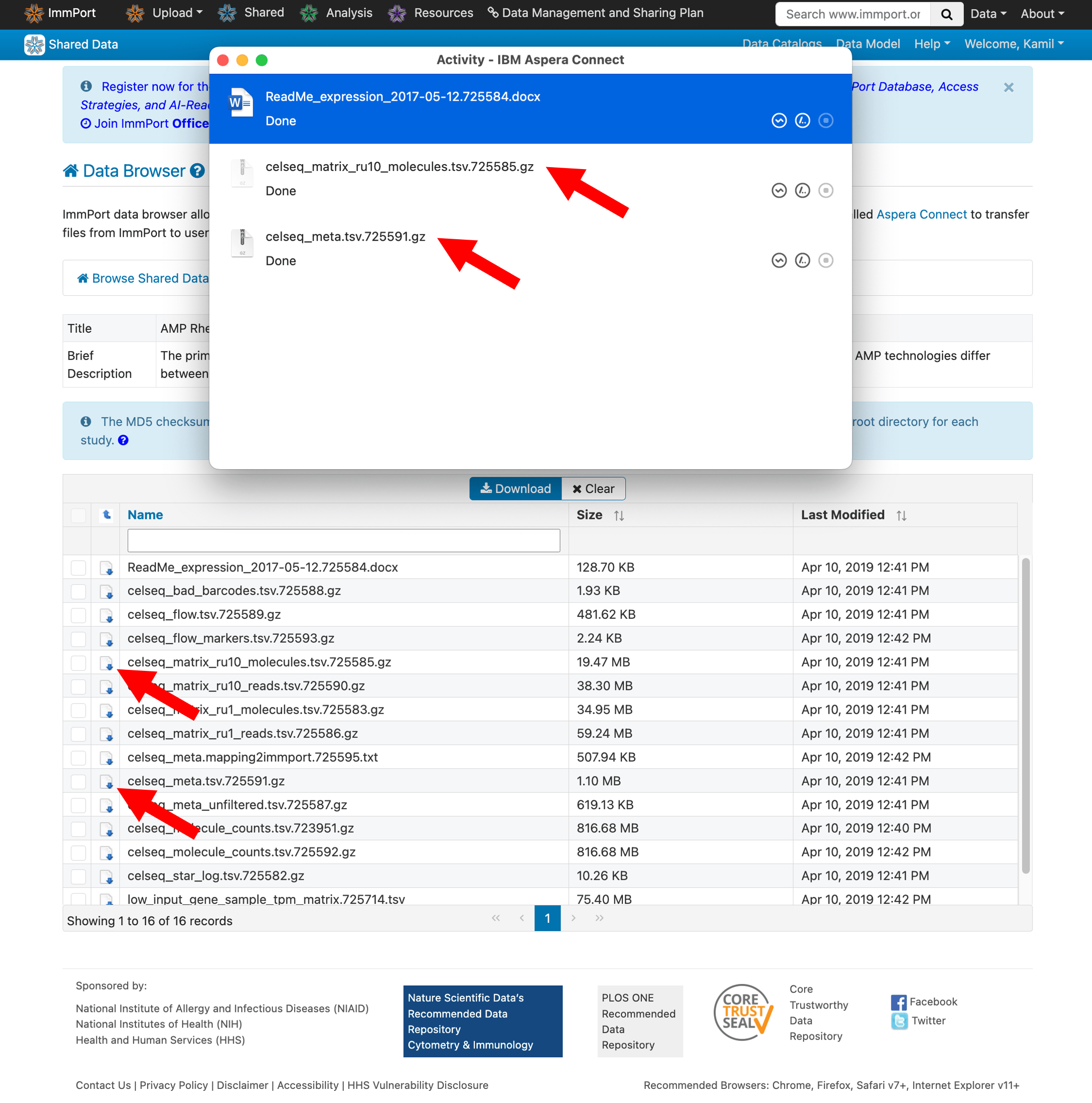
- Gene expression matrices (counts or TPM)
- Cell metadata and cluster annotations
- Sample metadata
Step 7: Load the data in R
Once downloaded, you can load the data files in R for analysis.
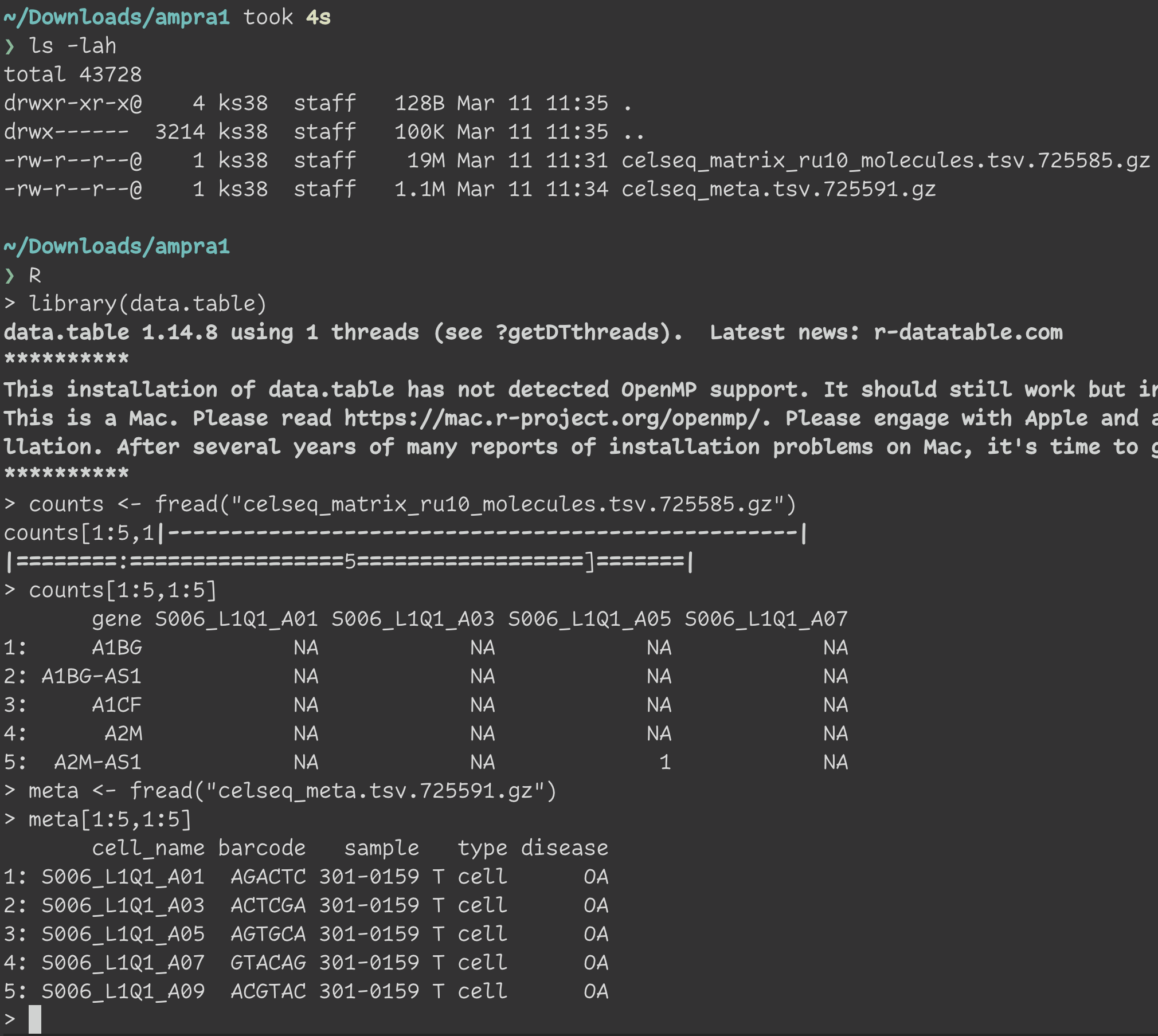
For detailed analysis examples, check out the amp_phase1_ra GitHub repository.
You're all set!
You now have access to the AMP RA Phase I single-cell RNA-seq data. If you use this data in your research, please cite:
Zhang F, Wei K, Slowikowski K, et al. Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-cell Transcriptomics and Mass Cytometry. Nature Immunology (2019). doi:10.1038/s41590-019-0378-1
Still need help?
If you run into any issues following this guide, feel free to reach out. We're happy to help.